Publications
(For a full list of Josh’s publications, including those prior to his time at MSU, go to Google Scholar, ORCID)
Highlights
Wicked cool simulations of a whole 11M atom BMC system! We learned alot from this research that we think we can apply to further advance catalysis within BMCs.
S Raza, NS Yadav, A Jussupow, CA Kerfeld, M Feig, JV Vermaas
J. Phys. Chem. B 129 12811-12827 (2025)
This particular research made the cover!
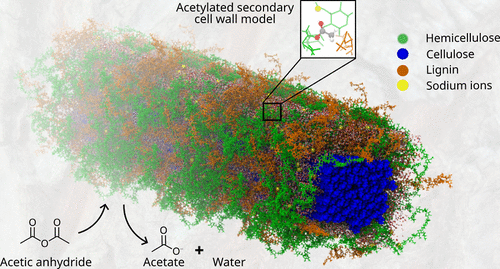
Complementary work tracking ion diffusion within plant cell walls after acetylation.
M Barkarar, D Sarkar, CG Hunt, JV Vermaas
Biomacromolecules 26 5645-5656 (2025)
This is a great example of undergraduate researchers doing cool work in the lab, as Murtaza has now moved on to bigger and better things!
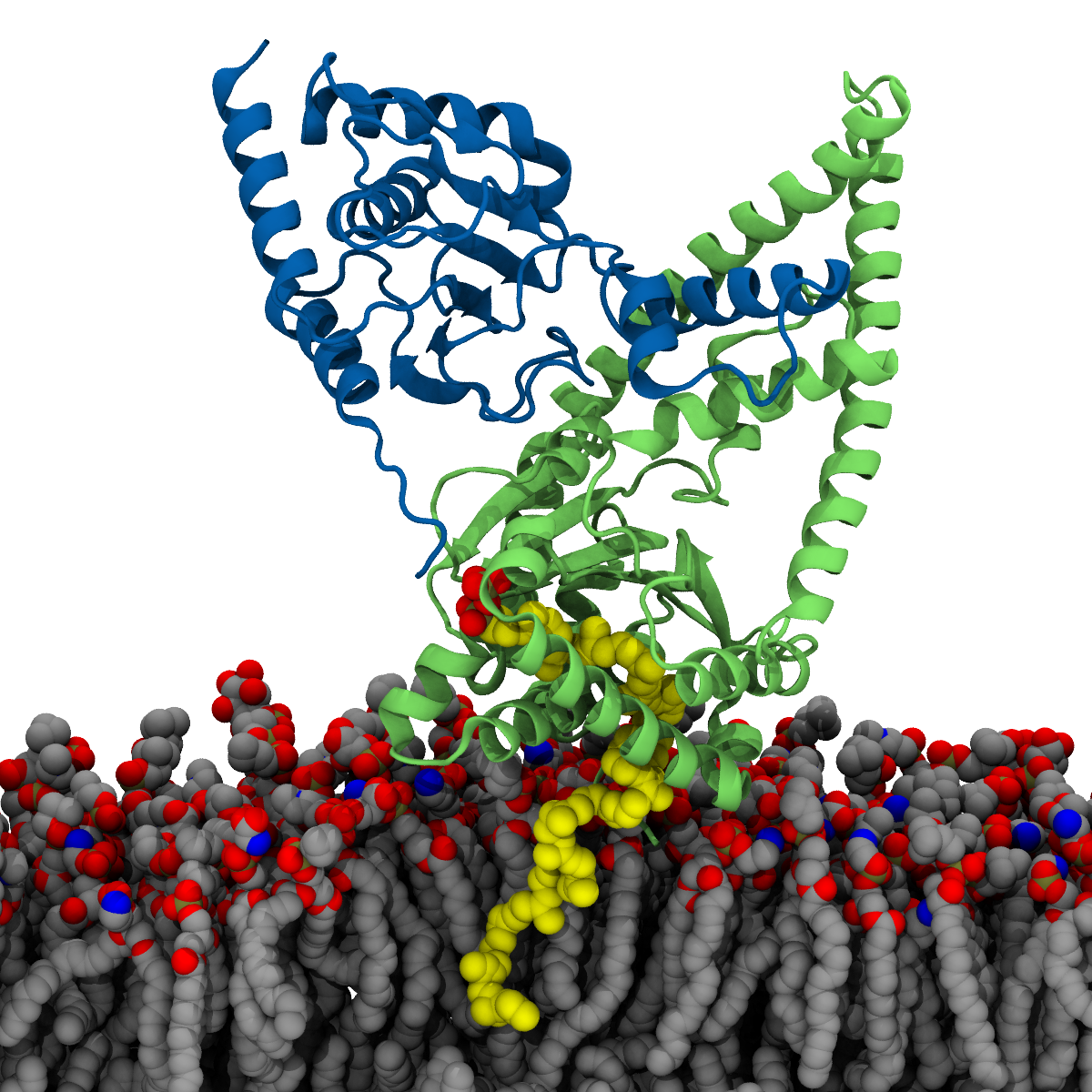
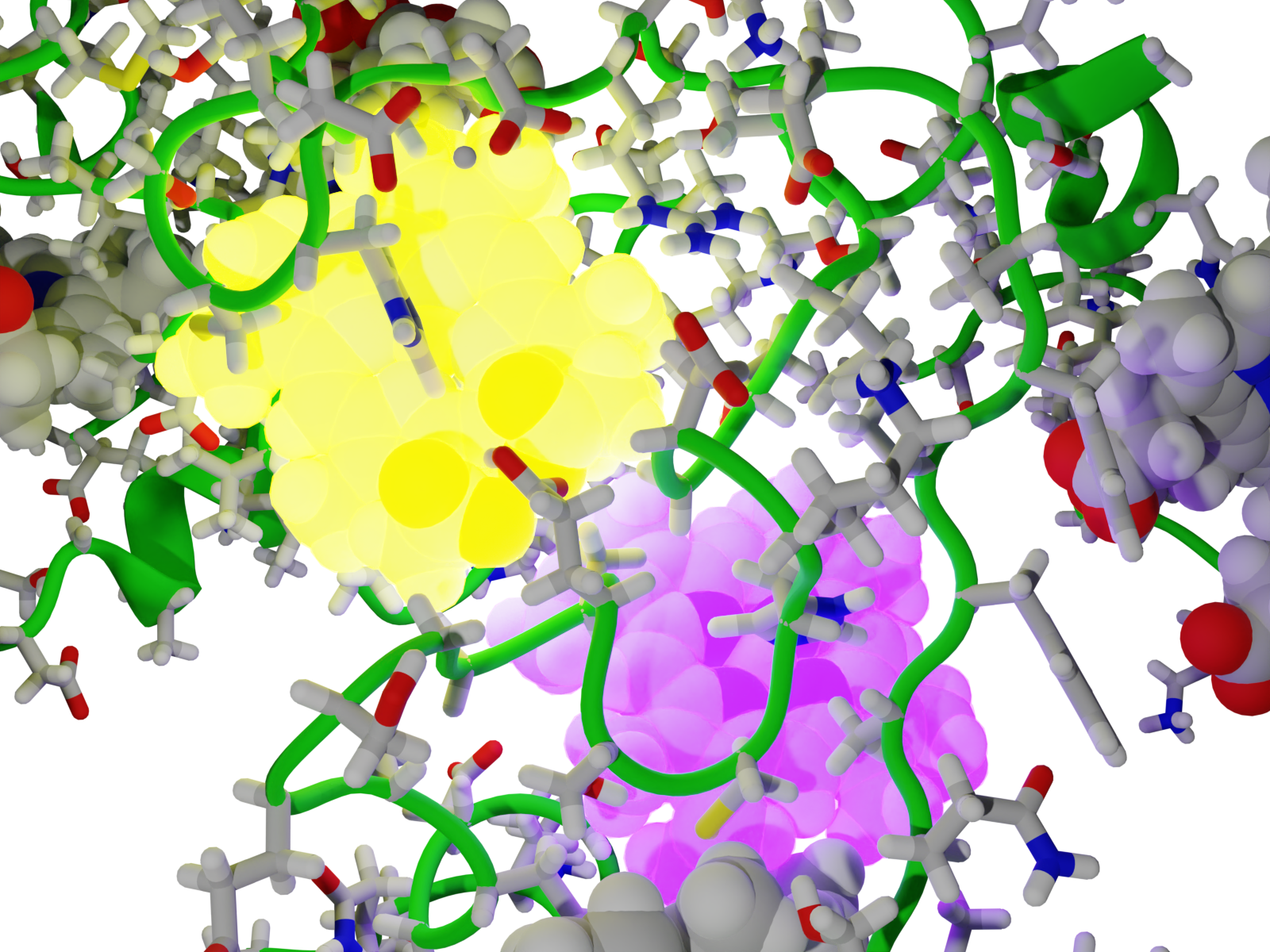
Bringing together simulation and experiment, we identify residues involved in protein binding for human cis-prenyltransferase to the membrane.
D Boren, S Kredi, E Positselskaya, M Giladi, Y Haitin, JV Vermaas
Protein Science 34 e70167 (2025)
This is the paper that Duncan won the Tomashow Keegstra Award for!
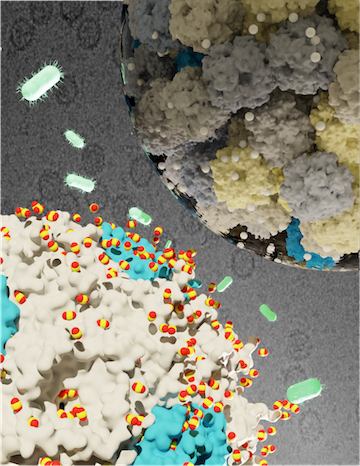
Through a combination of classical molecular dynamics simulations and atomic resolution Brownian dynamics, we determine the permeability for carboxysomes to photosynthetic metabolites and the rate of leakage from the carbon concentrating mechanism in cyanobacteria.
D Sarkar, C Maffeo, M Sutter, A Aksimentiev, CA Kerfeld, JV Vermaas
Kara did a fantastic news story on this work!
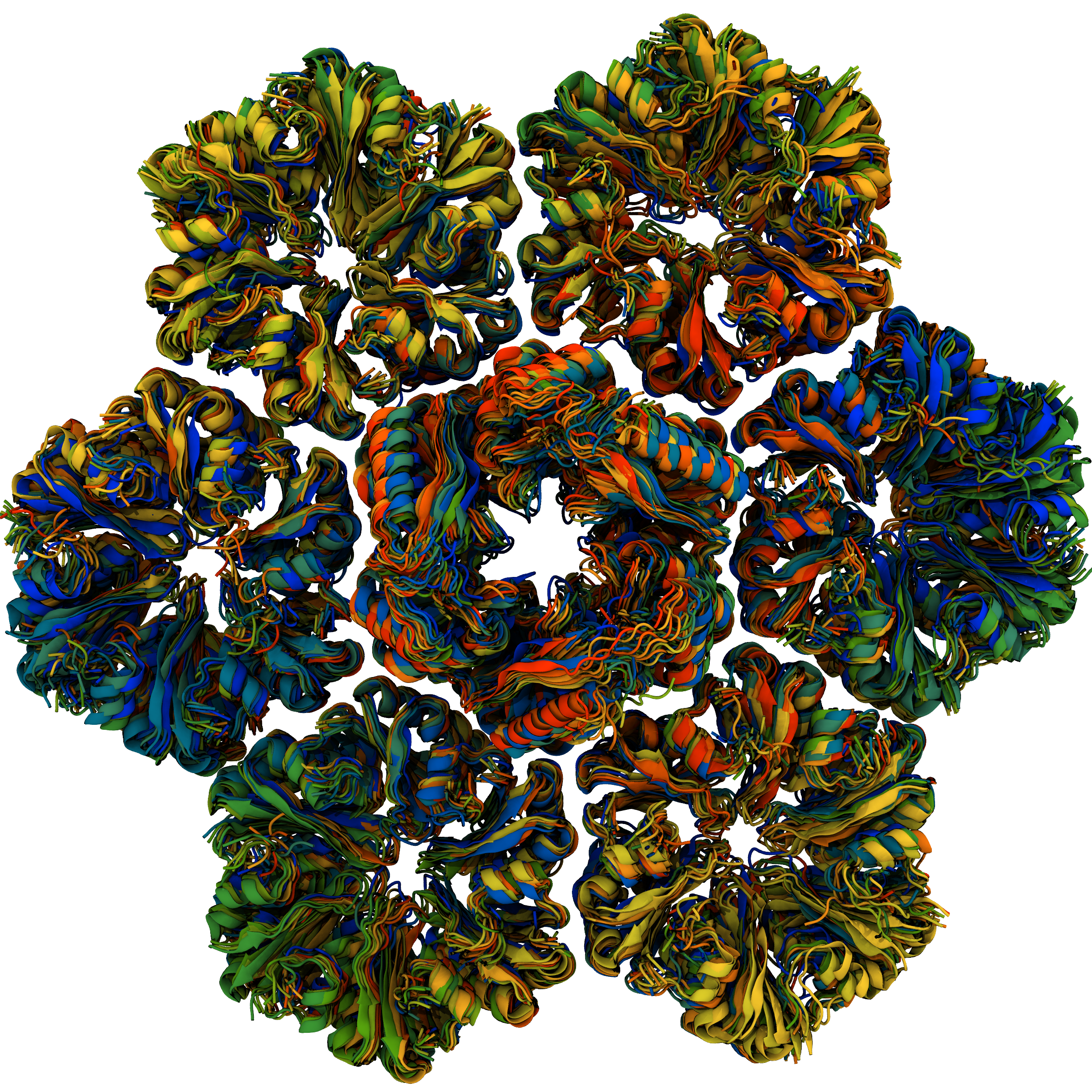
Comparing molecular simulation to x-ray footprinting mass spectrometry, we determine that flat and curved BMC protein assemblies have similar pore dynamics.
S Raza, D Sarkar, LJG Chan, J Mae, M Sutter, CJ Petzold, CA Kerfeld, CY Ralston, S Gupta, JV Vermaas
ACS Omega 9 35503-35514 (2024)
This paper was selected for a cover! We also have a news story to go along with the paper.
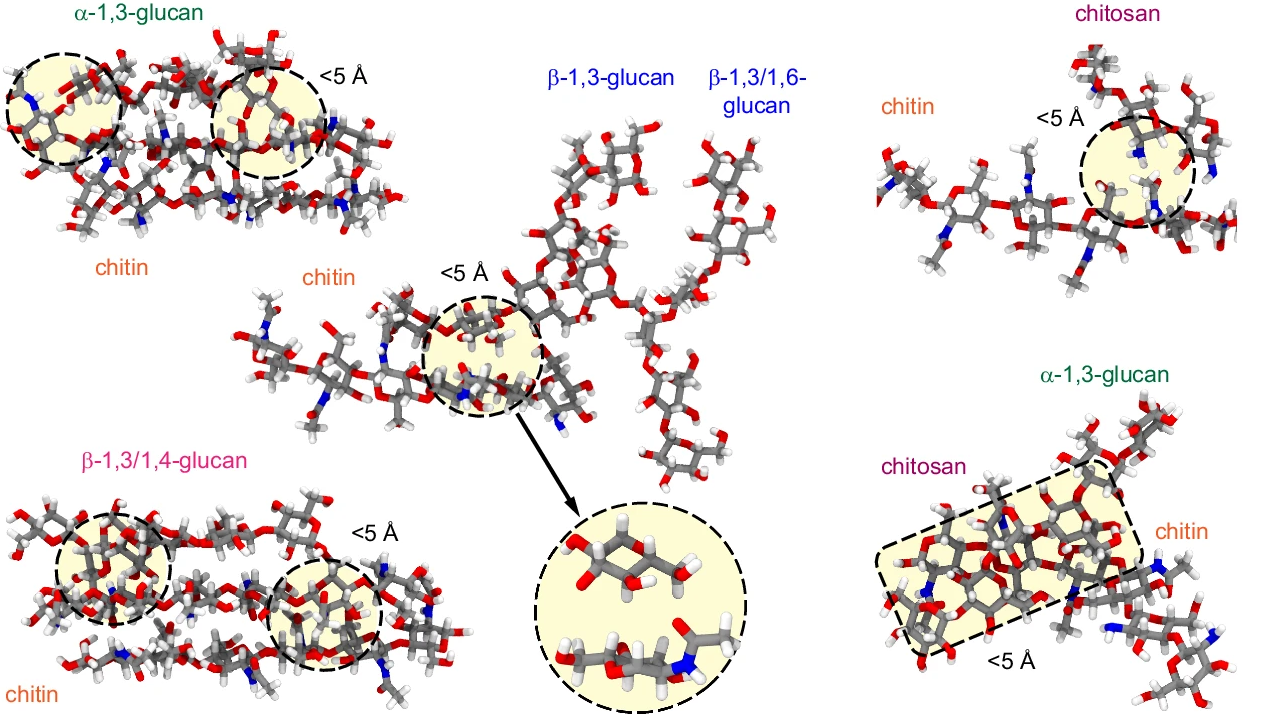
Using NMR together with molecular simulations, we can quantify how fungal cell walls change in response to drug exposure.
M Dickwella Widanage, I Gautam, D Sarkar, F Mentink-Vigier, JV Vermaas, SY Ding, AS Lipton, T Fontaine, JP Latgé, P Wang, and T Wang
Nature Communications 15 6382 (2024)
This paper has a high-level overview written by the MSU communications team!
Using higher level theory than prior papers, we get the order of magnitude but not the temperature dependence right for electron transfer within a nanocrystal.
W Parson, J Huang, M Kulke, JV Vermaas, and DM Kramer
Journal of Chemical Physics 160 065101 (2024)
This article was chosen for the journal cover, and the PRL has a great story about this research!
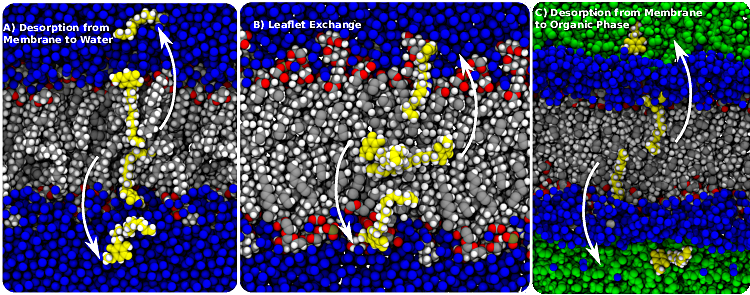
Based on molecular simulation, we find that sorgoleone, a small molecule made by sorghum, is readily able to transfer from cell to cell using passive membrane permeation.
S Raza, TH Sievertsen, S Okumoto, and JV Vermaas
Phytochemistry 217 113891 (2024)
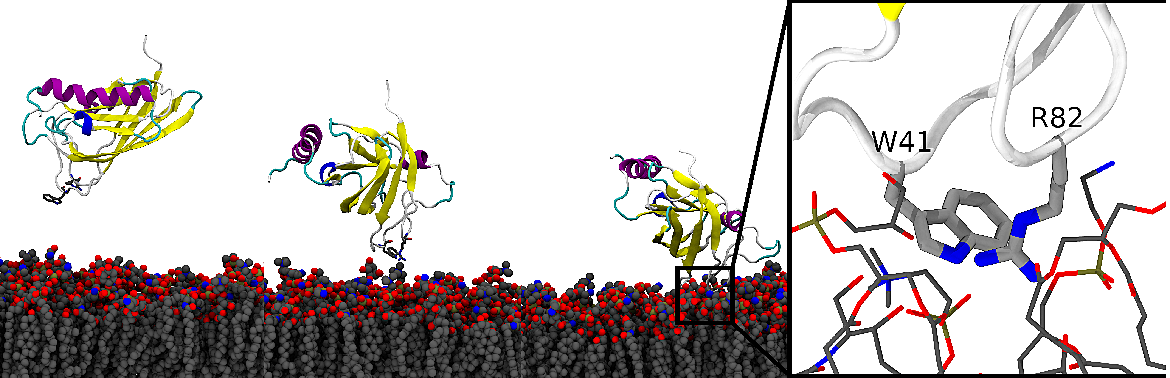
In this research article, we determine how PLAFP binds to biological membranes, and picks up a lipid, eventually transporting signaling lipids elsewhere in the cell.
M Kulke, E Kurtz, DM Boren, DM Olson, AM Koenig, S Hoffmann-Benning, and JV Vermaas
Plant Science 338 111900 (2024)
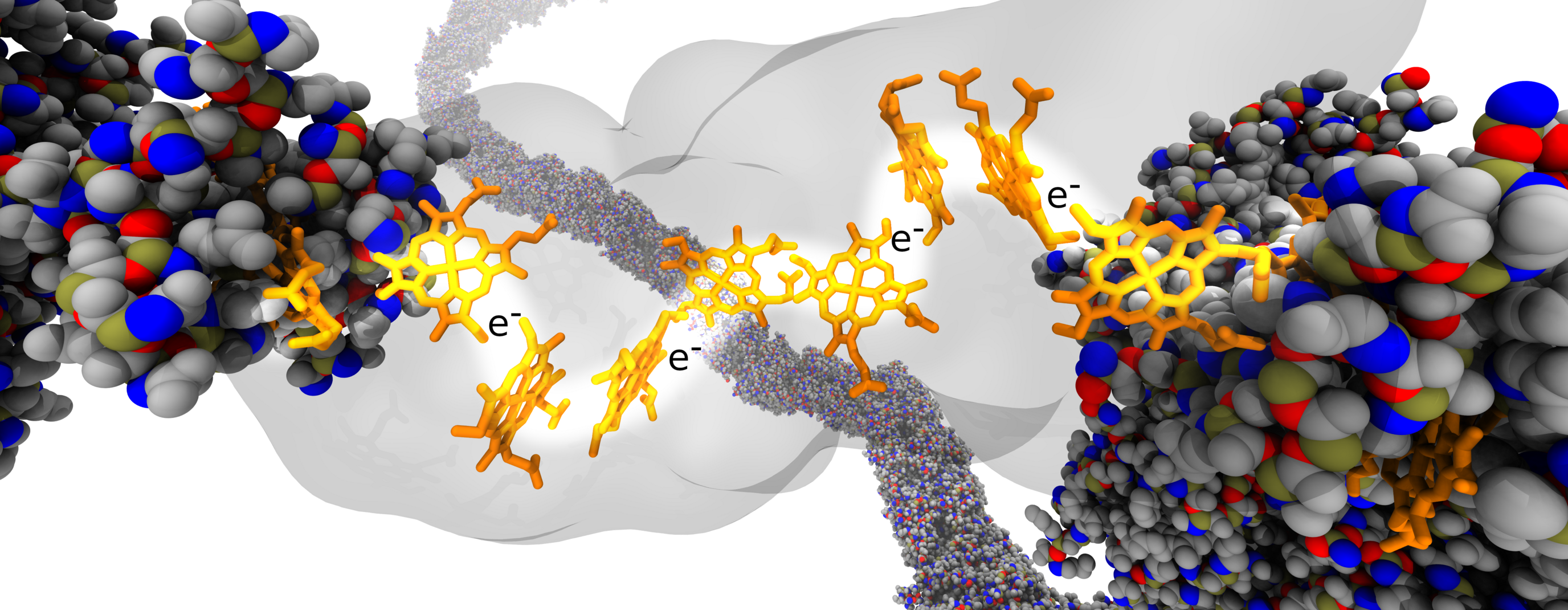
In this paper, we analyze how nanoscale vibrations in a cytochrome nanowire can influence the current carrying ability of these nanowires. Longer than normal distances that electrons need to “hop” create bottlenecks along large chains, which is ameliorated by having a wider wire for electron transfer to take place along.
M Kulke, DM Olson, J Huang, DM Kramer, and JV Vermaas
This research was picked up by MSU Today.

This work discusses the dynamic nature of the thylakoid membrane in response to temperature changes and its lipid composition. The study investigates the effects of temperature and isoprene content on the structure and dynamics of the membrane using molecular dynamics simulations. The results suggest that temperature affects various properties of the membrane, while isoprene concentration does not have a significant thermoprotective effect, and isoprene easily permeates the membrane.
M Kulke, SM Weraduwage, TD Sharkey, and JV Vermaas
Plant Cell & Environment 46 (8) 2273-2589 (2023)
This work was featured as a PRL news article.
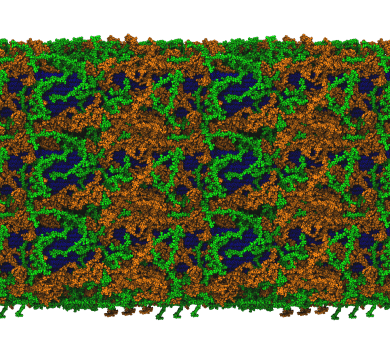
In collaboration with a diverse research group from NREL and the Forest Products Lab, we measured diffusion for ions and cell wall components within models for secondary cell walls at different hydration levels. Unsurprisingly, more water to lubricate the cell wall raises diffusion. However, we predict a moisutre-induced glass transition at around 10-15% water content in secondary plant cell walls.
D Sarkar, L Bu, JE Jakes, JK Zieba, ID Kaufman, MF Crowley, PN Ciesielski, and JV Vermaas
The Cell Surface 9 100105 (2023)
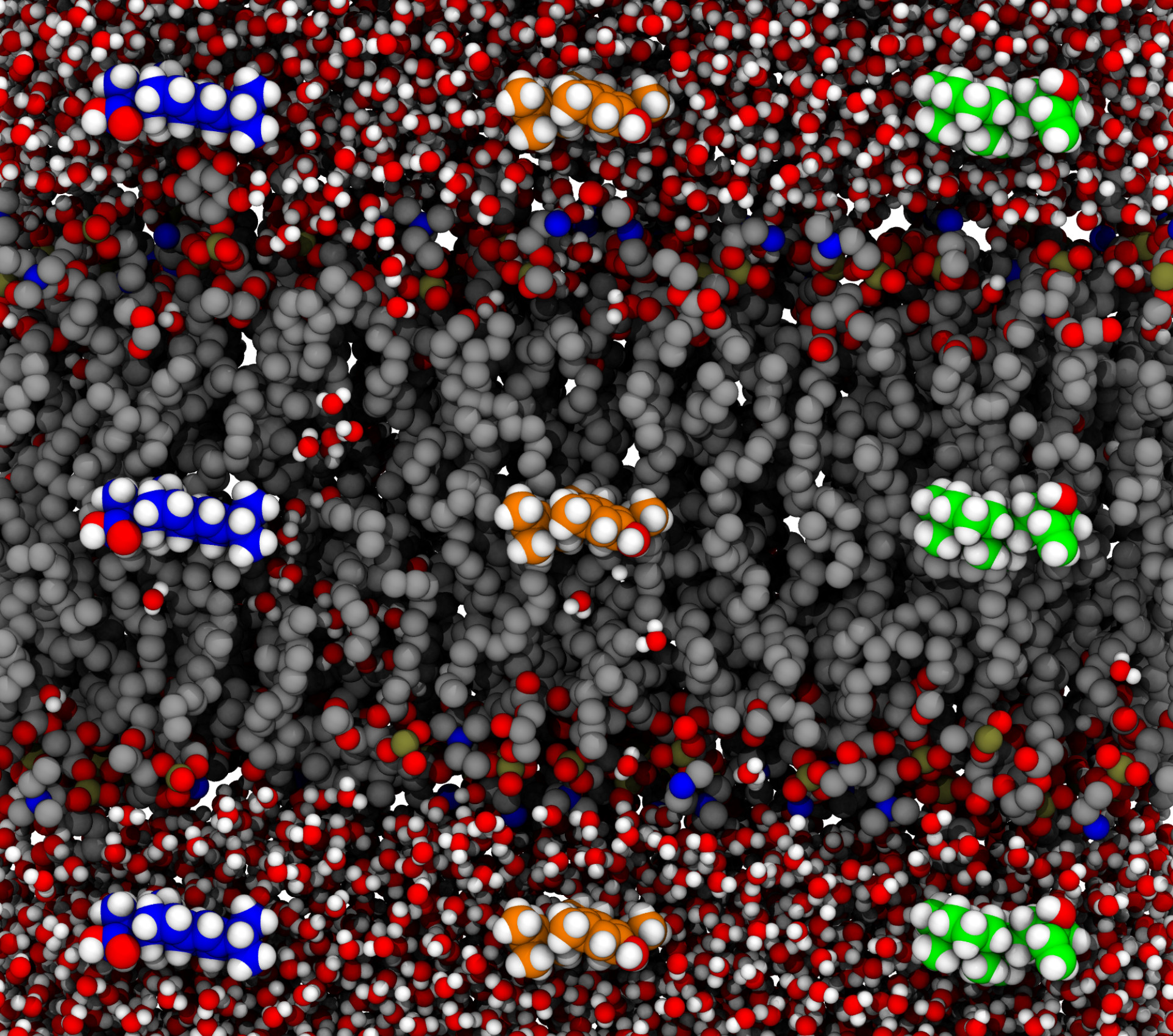
With this research, we explore how diterpenes permeate across biological membranes. We find that diterpenes permeate passively at large rates across bilayers. Intriguingly, we think that the permeation may be optimized for plant-like membranes.
S Raza, M Miller, B Hamberger, and JV Vermaas
Journal of Physical Chemistry B 127 (5) 1144-1157 (2023)
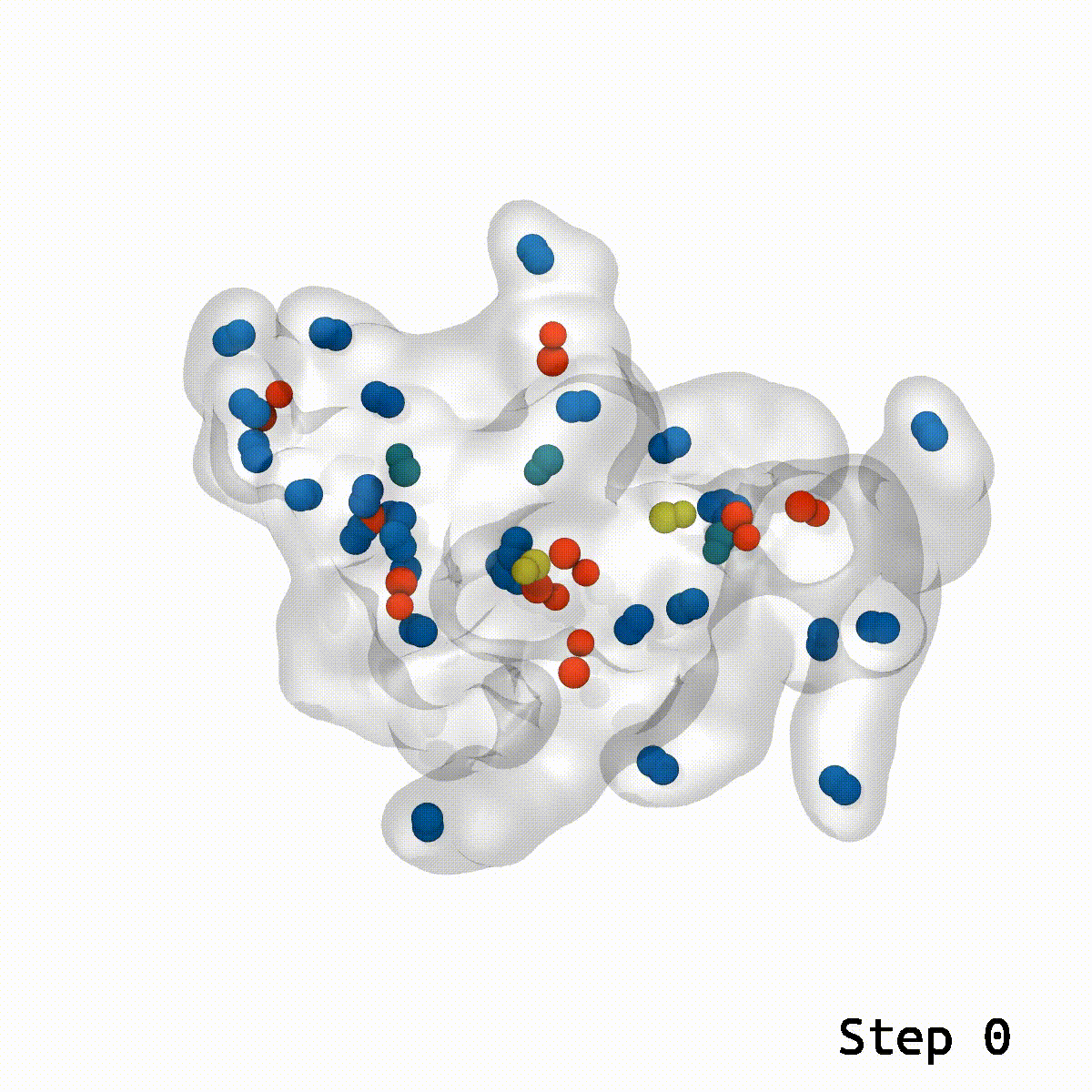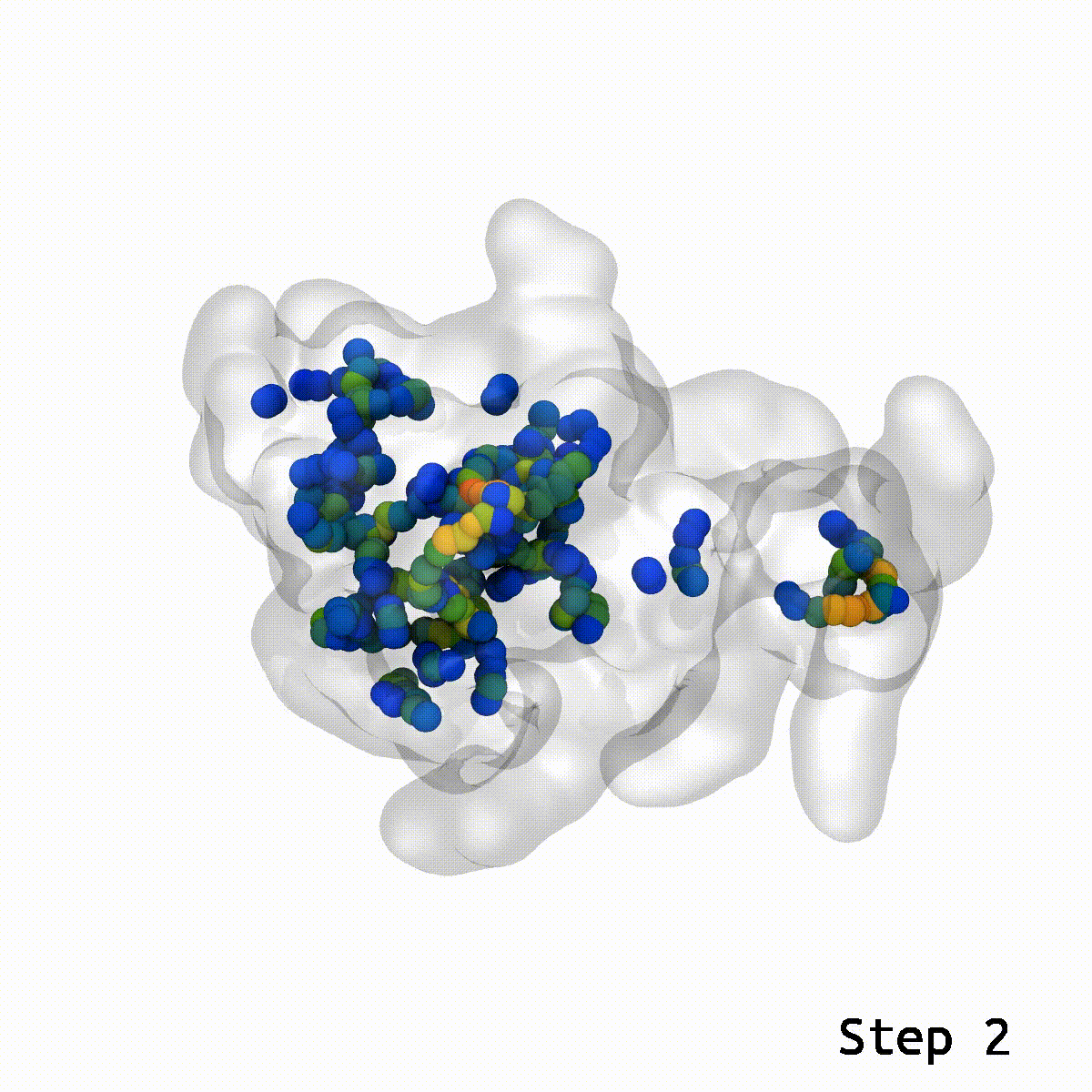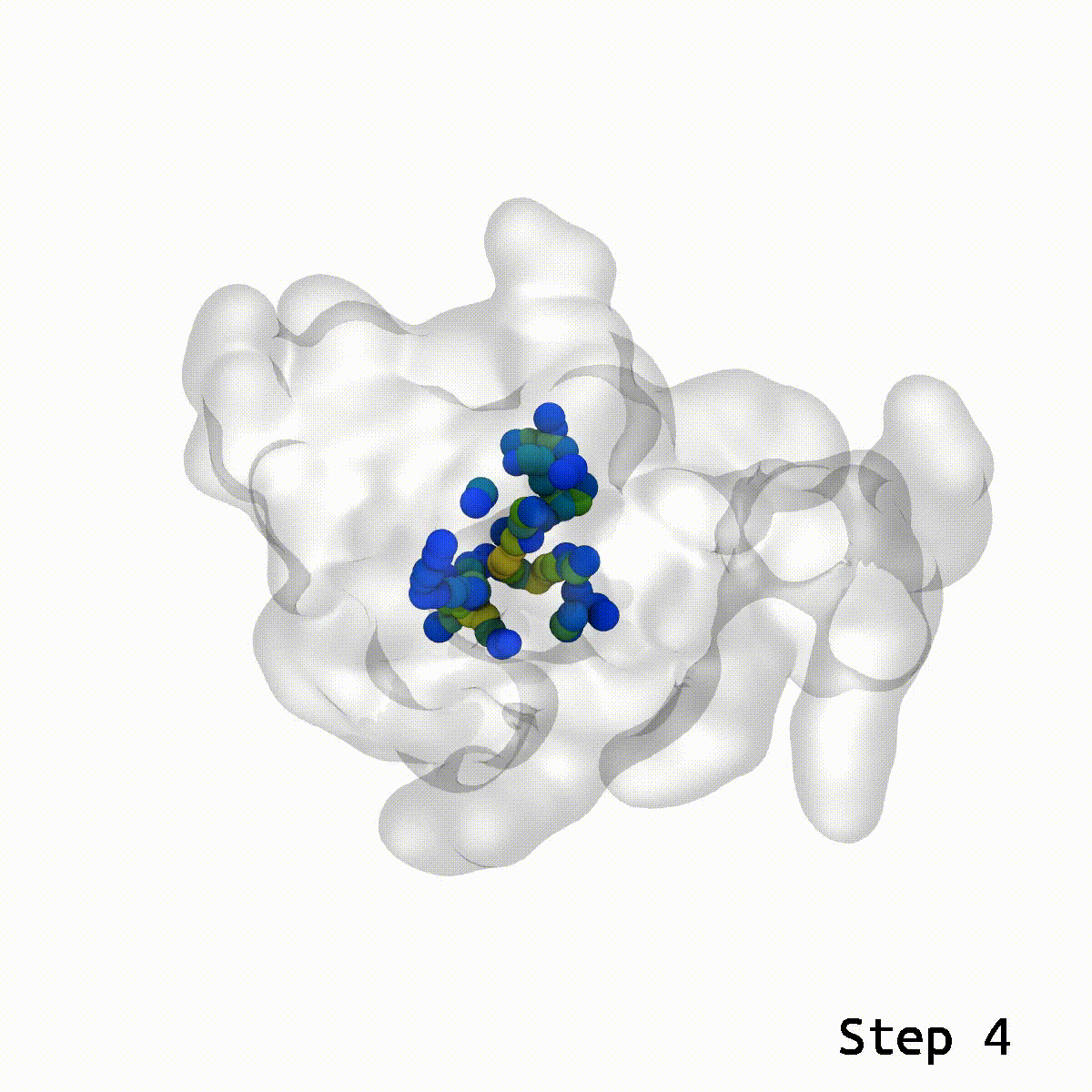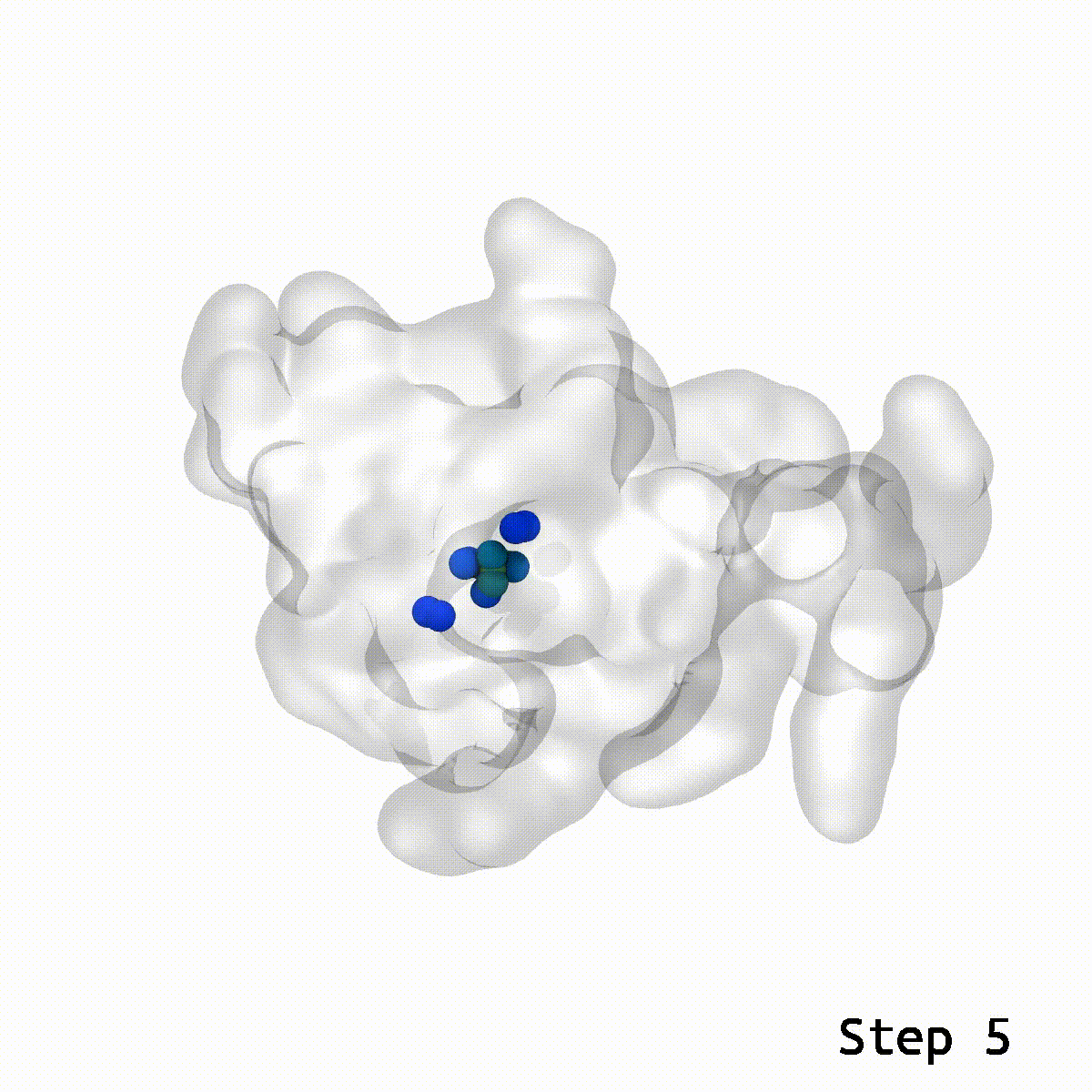
This is a cool tool to help relax away ring penetration artifacts that might arise when assembling a molecular simulation system.
D Sarkar, M Kulke, and JV Vermaas
Biomolecules 13 (1) 107 (2023)

Together with collaborators at MTU, we studied how lignin nanoparticles would form under different solvent conditions.
RCA Ela, S Raza, PA Heiden, JV Vermaas, and RG Ong
ACS Applied Polymer Materials 4 (10) 6925-6935 (2022)
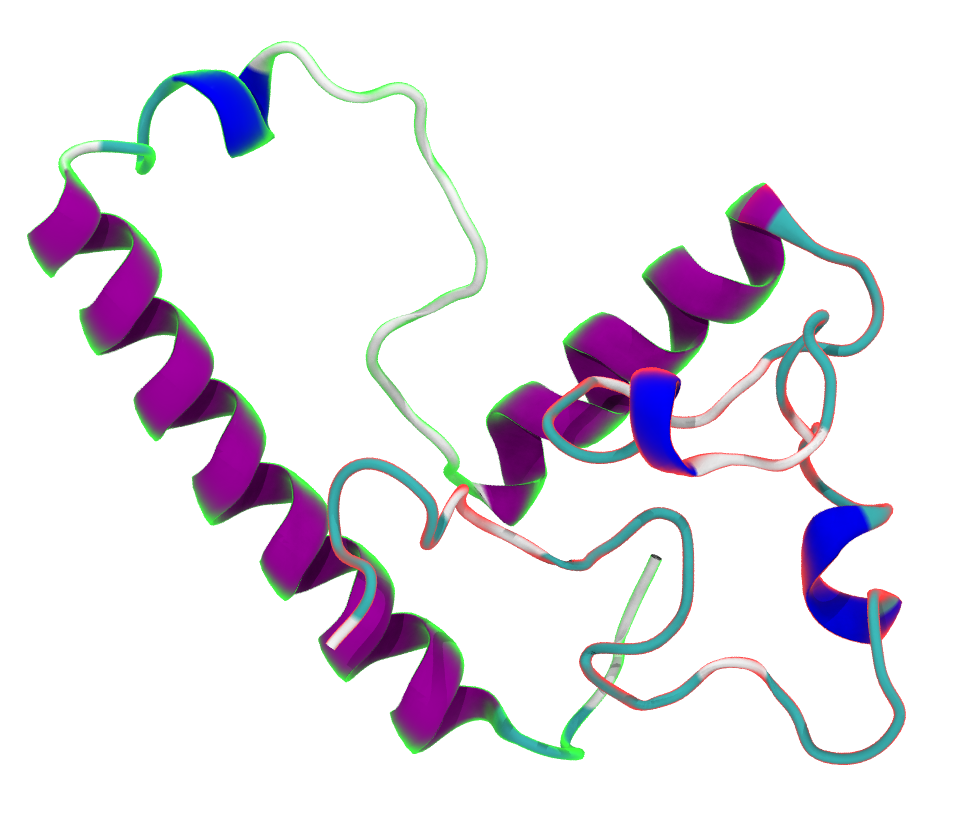
Together with collaborators in Germany, we refined a model for an intrinsicly disordered protein involved in Huntington’s disease.
SM Ayala Mariscal, ML Pigazzini, Y Richter, M Özel, IL Grothaus, J Protze, K Ziege, M Kulke, M ElBediwi, JV Vermaas, L Colombi Ciacchi, S Köppen, F Liu, and J Kirstein
Nature Communications 13, 4692 (2022)
Nature has a companion article that is less technical than a full-blown journal article.
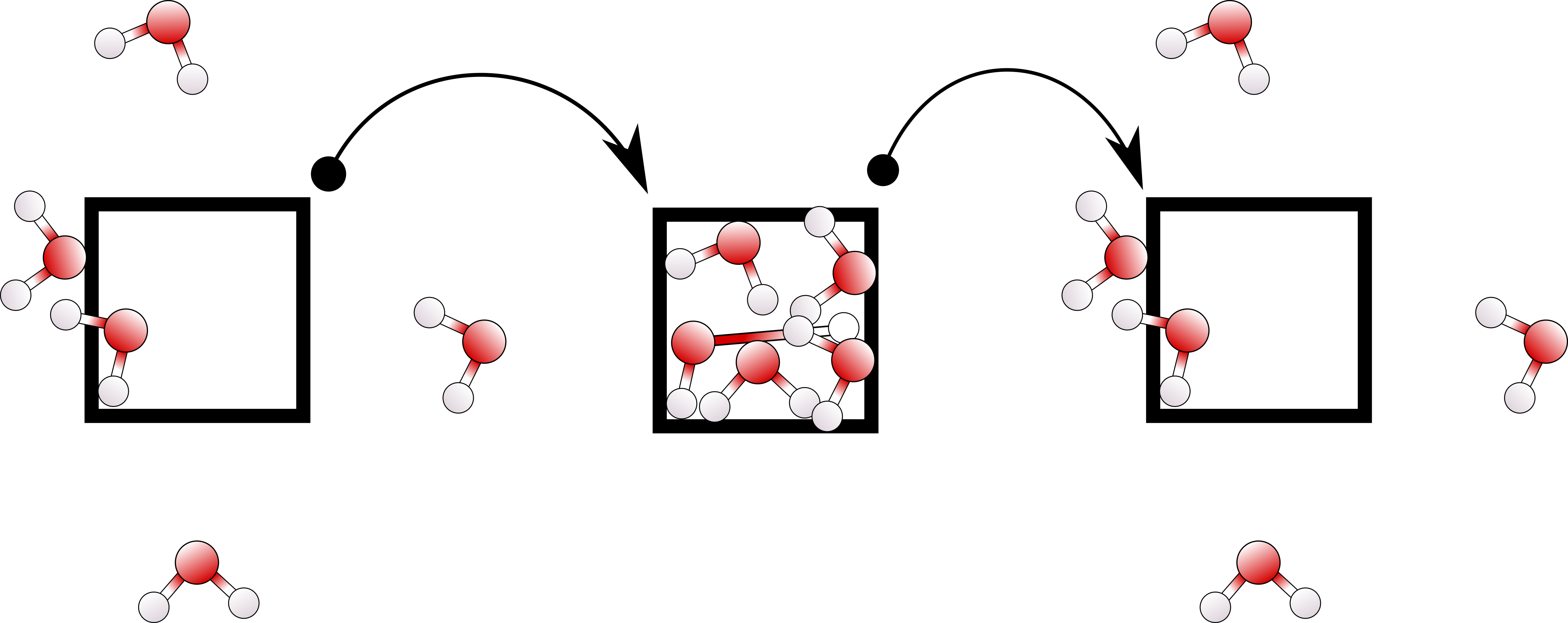
We developed a new scheme for doing PBC unwrapping, which is correct even for long simulations. We develop code implementations to apply this to qwrap, pbctools, and fastpbc.
M Kulke and JV Vermaas
Journal of Chemical Theory and Computation 18 (10) 6161-6171 (2022)
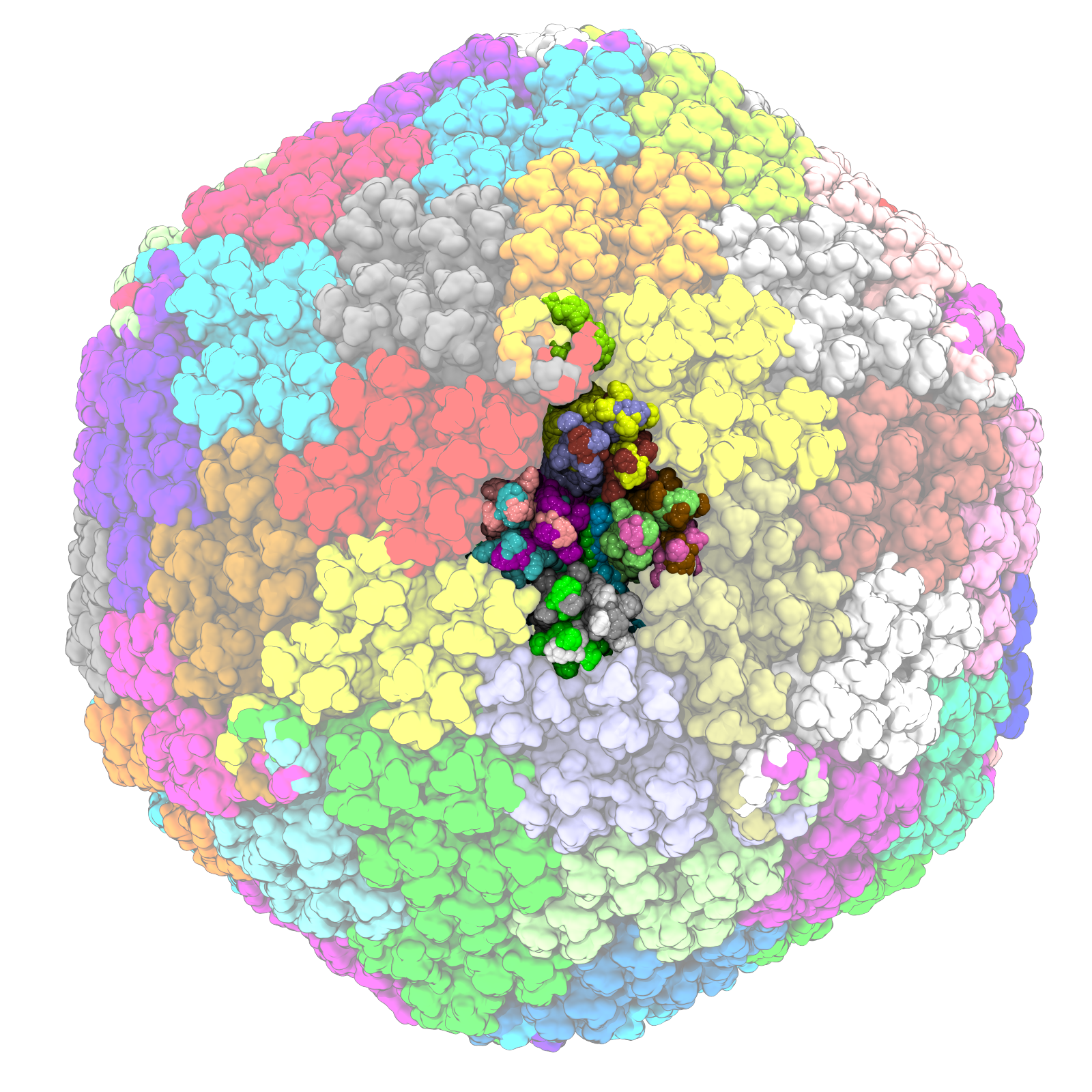
Using experimental Cryo-EM data, we built a better model for the viral vector used in the AstraZeneca COVID-19 vaccine. Our collaborators linked its structure to rare blood clotting side effects for the vaccine.
AT Baker, RJ Boyd, D Sarkar, A Teijeria-Crespo, CK Chan, E Bates, K Waraich, J Vant, E Wilson, CD Truong, M Lipka-Lloyd, P Fromme, J Vermaas, D Williams, L Machiesky, M Heurich, BM Nagalo, L Coughlan, S Umlauf, P-L Chui, PJ Rizkallah, TS Cohen, AL Parker, A Singharoy, and MJ Borad
This work has been featured by the BBC, EurekaAlert, and MSU.
Full List
Jojoba LIPID DROPLET-ASSOCIATED PROTEIN 1 facilitates the efficient packaging of wax esters into lipid droplets
P Whitehead, S Raza, M Miklaszewska, E Hornung, C Herrfurth, R Nadella, A Clews, NM Doner, JM Dyer, R Mullen, I Feussner, JV Vermaas, K Chapman
The Plant Cell 37 koaf115 (2025)
Molecular Modeling and Molecular Dynamics Simulation of a Packed and Intact Bacterial Microcompartment
S Raza, NS Yadav, A Jussupow, CA Kerfeld, M Feig, JV Vermaas
J. Phys. Chem. B 129 12811-12827 (2025)
Quantifying Chemically Modified Acetylation Induced Changes in the Plant Secondary Cell Wall Structure and Dynamics
M Barkarar, D Sarkar, CG Hunt, JV Vermaas
Biomacromolecules 26 5645-5656 (2025)
Anionic Lipids Regulate the Light-Harvesting Complex 1-Reaction Center Photocycle in Purple Bacteria
OC Fiebig, GP Schmidt, NS Yadav, D Wang, M Nairat, H Tang, Y Ji, V Prima, JN Sturgis, JV Vermaas, D Harris, GS Schlau-Cohen
JACS 147 36706-36716 (2025)
Identifying and quantifying membrane interactions of the protein human cis‐prenyltransferase
D Boren, S Kredi, E Positselskaya, M Giladi, Y Haitin, JV Vermaas
Protein Science 34 e70167 (2025)
Synthesis, function, and genetic variation of sorgoleone, the major biological nitrification inhibitor in sorghum
S Okumoto, B Maharjan, N Rajan, J Xi, SR Baerson, WL Rooney, MJ Tomson, DA Odeny, T Yoshihashi, JV Vermaas, GV Subbarao
Quantitative Measurement of Molecular Permeability to a Synthetic Bacterial Microcompartment Shell System
EJ Young, H Kirst, ME Dwyer, JV Vermaas, CA Kerfeld
ACS Synth. Biol. 14 1405-1413 (2025)
Structure Characterization of Bacterial Microcompartment Shells via X-ray Scattering and Coordinate Modeling: Evidence for Adventitious Capture of Cytoplasmic Proteins
X Zuo, A Jussupow, NS Ponomarenko, TD Grant, TM Tefft, NS Yadav, KL Range, CY Ralston, MA TerAvest, M Sutter, CA Kerfeld, JV Vermaas, M Feig, DM Tiede
ACS Appl. Bio Mater. 8 2090-2103 (2025)
Electrochemical cofactor recycling of bacterial microcompartments
M Sutter, LM Utschig, J Niklas, S Paul, DN Kahan, S Gupta, OG Poluektov, BH Ferlez, NM Tefft, MA TerAvest, DP Hickey, JV Vermaas, CY Ralston, CA Kerfeld
PNAS 121 e2414220121 (2024)
Increasing thermostability of the key photorespiratory enzyme glycerate 3-kinase by structure-based recombination
LV Roze, A Antoniak, D Sarkar, AH Liepman, M Tejera-Nieves, JV Vermaas, BJ Walker
Plant Biotechnology Journal 23 454-466 (2025)
Atomic View of Photosynthetic Metabolite Permeability Pathways and Confinement in Synthetic Carboxysome Shells
D Sarkar, C Maffeo, M Sutter, A Aksimentiev, CA Kerfeld, JV Vermaas
PNAS 121 e2402277121 (2024)
Comparative Pore Structure and Dynamics for Bacterial Microcompartment Shell Protein Assemblies in Sheets or Shells
S Raza, D Sarkar, LJG Chan, J Mae, M Sutter, CJ Petzold, CA Kerfeld, CY Ralston, S Gupta, JV Vermaas
ACS Omega 9 35503-35514 (2024)
Adaptative survival of Aspergillus fumigatus to echinocandins arises from cell wall remodeling beyond β−1,3-glucan synthesis inhibition
M Dickwella Widanage, I Gautam, D Sarkar, F Mentink-Vigier, JV Vermaas, SY Ding, AS Lipton, T Fontaine, JP Latgé, P Wang, and T Wang
Nature Communications 15 6382 (2024)
The isoprene‐responsive phosphoproteome provides new insights into the putative signalling pathways and novel roles of isoprene
SM Weraduwage, D Whitten, M Kulke, A Sahu, JV Vermaas, and TD Sharkey
Plant, Cell & Environment 47 1099-1117 (2024)
Electron transfer in a crystalline cytochrome with four hemes
W Parson, J Huang, M Kulke, JV Vermaas, and DM Kramer
Journal of Chemical Physics 160 065101 (2024)
Passive permeability controls synthesis for the allelochemical sorgoleone in sorghum root exudate
S Raza, TH Sievertsen, S Okumoto, and JV Vermaas
Phytochemistry 217 113891 (2024)
PLAT domain protein 1 (PLAT1/PLAFP) binds to the Arabidopsis thaliana plasma membrane and inserts a lipid
M Kulke, E Kurtz, DM Boren, DM Olson, AM Koenig, S Hoffmann-Benning, and JV Vermaas
Plant Science 338 111900 (2024)
Adaptive Ensemble Refinement of Protein Structures in High Resolution Electron Microscopy Density Maps with Radical Augmented Molecular Dynamics Flexible Fitting
D Sarkar, H Lee, JW Vant, M Turilli, JV Vermaas, S Jha, and A Singharoy
Journal of Chemical Information and Modeling 63 (18) 5834-5846 (2023)
Long-Range Electron Transport Rates Depend on Wire Dimensions in Cytochrome Nanowires
M Kulke, DM Olson, J Huang, DM Kramer, and JV Vermaas
Small 19 (52) 2304013 (2023)
Nanoscale simulation of the thylakoid membrane response to extreme temperatures
M Kulke, SM Weraduwage, TD Sharkey, and JV Vermaas
Plant Cell & Environment 46 (8) 2273-2589 (2023)
Diffusion in intact secondary cell wall models of plants at different equilibrium moisture content
D Sarkar, L Bu, JE Jakes, JK Zieba, ID Kaufman, MF Crowley, PN Ciesielski, and JV Vermaas
The Cell Surface 9 100105 (2023)
Atomistic origins of biomass recalcitrance in organosolv pretreatment
D Sarkar, IJ Santiago, and JV Vermaas
Chemical Engineering Science 272 118587 (2023)
Characterization of promoter elements of isoprene‐responsive genes and the ability of isoprene to bind START domain transcription factors
SM Weraduwage, A Sahu, M Kulke, JV Vermaas, and TD Sharkey
Plant Direct 7 (2) e483 (2023)
Plant Terpenoid Permeability through Biological Membranes Explored via Molecular Simulations
S Raza, M Miller, B Hamberger, and JV Vermaas
Journal of Physical Chemistry B 127 (5) 1144-1157 (2023)
LongBondEliminator: A Molecular Simulation Tool to Remove Ring Penetrations in Biomolecular Simulation Systems
D Sarkar, M Kulke, and JV Vermaas
Biomolecules 13 (1) 107 (2023)
Lignin Nanoparticle Morphology Depends on Polymer Properties and Solvent Composition: an Experimental and Computational Study
RCA Ela, S Raza, PA Heiden, JV Vermaas, and RG Ong
ACS Applied Polymer Materials 4 (10) 6925-6935 (2022)
Identification of a HTT-specific binding motif in DNAJB1 essential for suppression and disaggregation of HTT
SM Ayala Mariscal, ML Pigazzini, Y Richter, M Özel, IL Grothaus, J Protze, K Ziege, M Kulke, M ElBediwi, JV Vermaas, L Colombi Ciacchi, S Köppen, F Liu, and J Kirstein
Nature Communications 13, 4692 (2022)
Exploring cryo-electron microscopy with molecular dynamics
JW Vant, D Sarkar, J Nguyen, AT Baker, JV Vermaas, A Singharoy
Biochemical Society Transactions 50 (1) 569-581 (2022)
Investigating Reversibility in Unwrapping Schemes for Molecular Dynamics Simulations
M Kulke and JV Vermaas
Journal of Chemical Theory and Computation 18 (10) 6161-6171 (2022)
ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome
AT Baker, RJ Boyd, D Sarkar, A Teijeria-Crespo, CK Chan, E Bates, K Waraich, J Vant, E Wilson, CD Truong, M Lipka-Lloyd, P Fromme, J Vermaas, D Williams, L Machiesky, M Heurich, BM Nagalo, L Coughlan, S Umlauf, P-L Chui, PJ Rizkallah, TS Cohen, AL Parker, A Singharoy, and MJ Borad
Science Advances 7, 49 (2021)