Research
The overall goal for the group is to use computation to develop models to answer pivotal questions around plant materials and photosynthetic processes. Specifically, we use molecular simulation to focus on atomic-scale interactions that govern plant processes, generating testable hypotheses that can be tested on the bench or in a field by our collaborators.
Here are some themes in our current research:
Lipid membranes 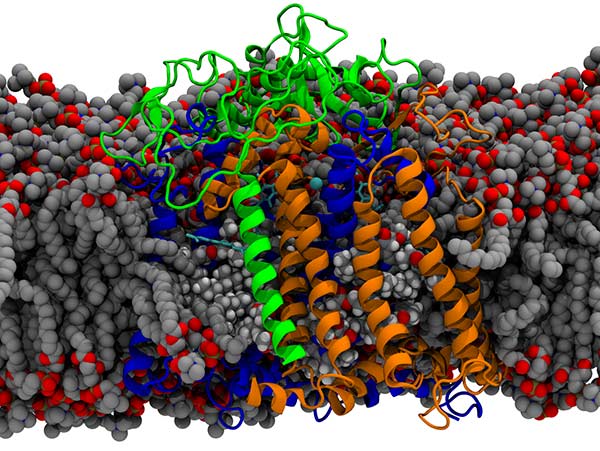 Lipid bilayers are the fundamental biological structure that partitions cellular structures, creating gradients across the lipid leaflets that drive cellular metabolism and transport processes.
Our interest is in how membranes influence protein function, such as by binding to specific sites on proteins as above, as well as influence metabolic fluxes within cells by controlling transport.
To do this, we create atomic models using classical molecular simulation approaches, and carry out biased and unbiased molecular simulations to answer our specific molecular hypotheses.
Lipid bilayers are the fundamental biological structure that partitions cellular structures, creating gradients across the lipid leaflets that drive cellular metabolism and transport processes.
Our interest is in how membranes influence protein function, such as by binding to specific sites on proteins as above, as well as influence metabolic fluxes within cells by controlling transport.
To do this, we create atomic models using classical molecular simulation approaches, and carry out biased and unbiased molecular simulations to answer our specific molecular hypotheses.
Cellular structures and organelles
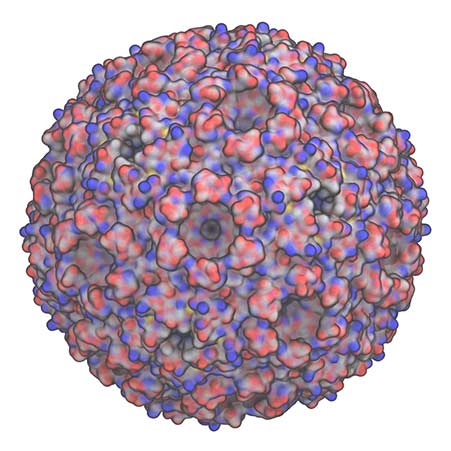 Similarly, we want to understand how structure and function are coupled together across biology, such as in the carboxysome shown here, or in larger biomass assemblies.
This will allow for these systems to be engineered, either for enhanced degradation into industrial products, or to repurpose the system to generate alternative products.
Similarly, we want to understand how structure and function are coupled together across biology, such as in the carboxysome shown here, or in larger biomass assemblies.
This will allow for these systems to be engineered, either for enhanced degradation into industrial products, or to repurpose the system to generate alternative products.
New simulation methods
 Underlying these structural studies, we also want to advance new simulation techniques and incorporate new simulation methods into our research.
One such example is by using machine-learned force fields to break apart lignin, which might be applicable to catalysis in other systems.
Underlying these structural studies, we also want to advance new simulation techniques and incorporate new simulation methods into our research.
One such example is by using machine-learned force fields to break apart lignin, which might be applicable to catalysis in other systems.